Evotec offers end-to-end membrane protein services through a dedicated expert platform. With proven success in GPCRs, ion channels, solute carriers, and other challenging targets, we deliver high-quality proteins optimized for structural biology and biophysical analysis.
From Gene Design to Structural Readiness
Evotec’s membrane protein biochemistry expertise enables optimized gene and construct design for high-yield expression across challenging targets. Our automated platform with KingFisher APEXTM accelerates expression scouting and detergent screening to produce highly pure, monodisperse proteins ready for biophysical assays, structural biology, or reconstitution into lipid nanodiscs and polymer systems.
Evotec's Expertise
Whether your focus is on GPCRs, ion channels, or transporters, Evotec combines deep scientific expertise with advanced technologies to accelerate discovery and development.
- Custom expression systems (mammalian: transient & BacMam, insect)
- Detergent-based and native-like solubilization (lipid nanodiscs, copolymers)
- High-throughput screening and protein stabilization
- Biophysical and structural characterization
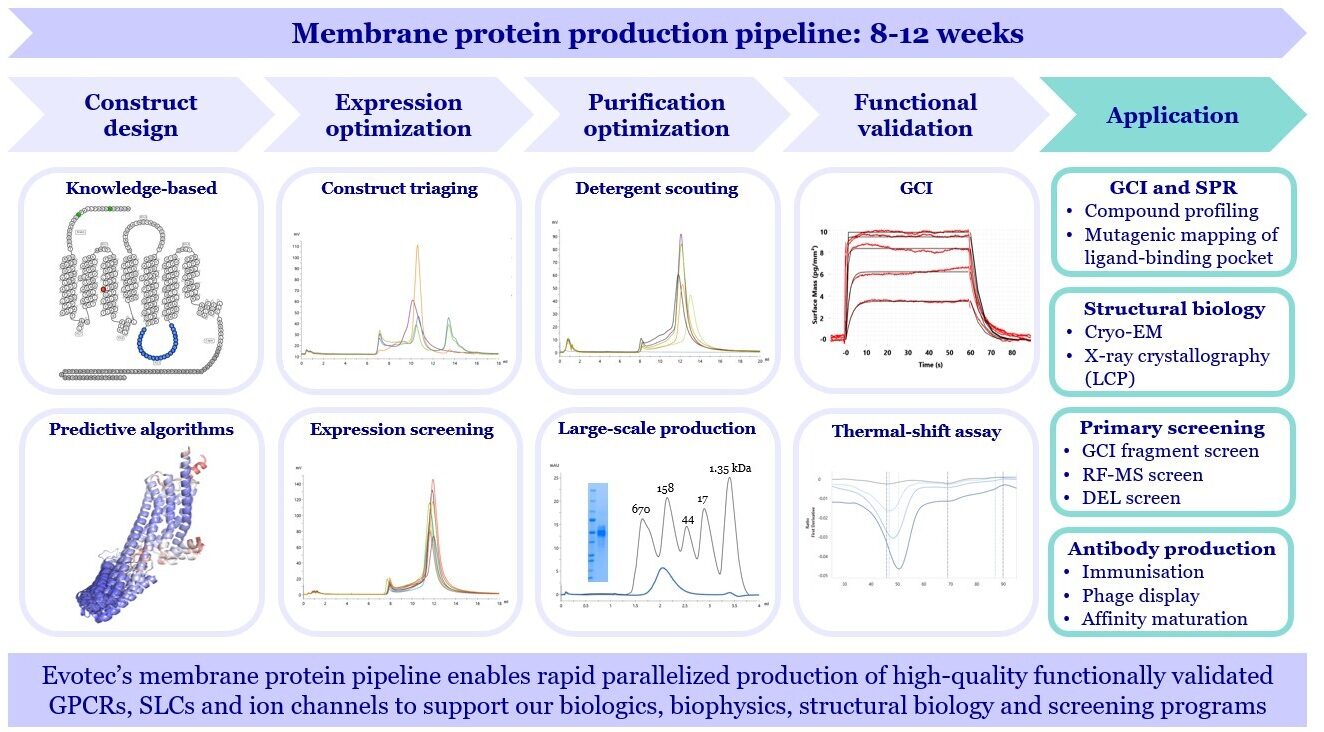
Membrane Protein Pipeline: From Construct Design to Functional Validation in 8–12 Weeks
Why Evotec?
- Proven success with high-value, difficult-to-express targets
- Fast turnaround and scalable production
- Support across discovery, preclinical, and structural biology workflows
Frequently Asked Questions
Q: What is the optimal size of a membrane protein for cryo-EM at Evotec?
For high-resolution cryo-EM structure determination, membrane proteins should ideally be >100 kDa and free of flexible or disordered regions. For smaller or more dynamic targets, Evotec designs tailored strategies such as adding binder proteins or stabilizing flexible regions to enable reliable structure determination.
Q: What resolution can I expect from a cryo-EM structure of a membrane protein?
With recent advances in cryo-EM services, Evotec routinely achieves resolutions as high as 2 Å. For most membrane proteins, density maps typically reach 2.5–3 Å, sufficient to define the ligand-binding pose. Focused data processing can further enhance local resolution at binding sites or other regions of interest.
Q: What is the throughput of GCI for compound screening against membrane proteins?
Using the waveRAPID injection method, up to 352 fragments can be screened per day, or 96 compounds tested against three targets in a single experiment. Once promising binders are identified, multicycle kinetics provide detailed steady-state kinetics and final concentration-response curves (CRC).