Evotec strongly believes in the power of omics-driven drug discovery. The unbiased nature and magnitude of insight enabled by multi-omics will allow us to tackle the challenges ahead. Healthcare and optimal medical treatment of patients will require more sophisticated, higher efficient and highly selective drugs.
If we want to uncover the full complexity of diseases, we must understand the underlying molecular processes driving them and decipher the reasons for their various phenotypes in a diverse range of patients. This will pave the way for the development of new and precise cures and how to better identify the correct patients to treat. Today, we can observe and explore the processes on a molecular level using unbiased genomics, transcriptomics, proteomics, and metabolomics to provide unprecedented insights. Each layer of understanding alone is already a milestone towards a more general understanding. Interconnecting these datasets in a multi-omics analysis, together with relevant clinical data, however, will reveal the whole spectrum of influences on the intracellular processes by diseases and treatments.
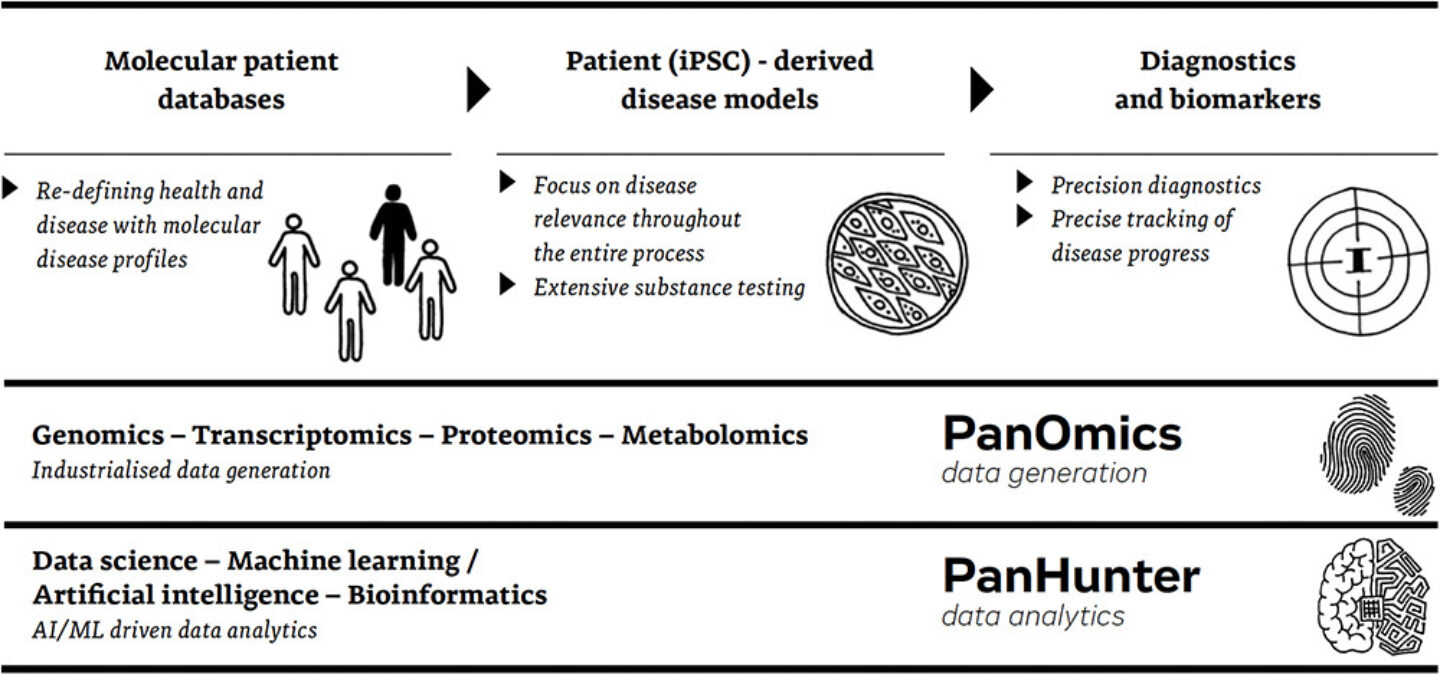
Figure 1: Evotec’s omics-driven and patient-based research platforms
This drive for deep understanding motivates all steps we do, every step is therefore backed-up by extensive data collection and analysis, and we are in a great position for this approach as we can combine the various strengths we built-up with our partners over the last decade. From molecular patient databases, we derive disease profiles that we translate into – for example – iPSC-enabled disease models. Against those models, we can develop and screen very large amounts of compounds and really utilize the industry leading high throughput platforms we have created in our panomics projects to generate the multi-omics data we need.
However, generating deep and high quality omics data is just the starting point. Alongside advancing our data generation and interpretation capabilities, we have been working towards democratizing access to data exploration for all scientists and give all researchers the bio – informatics skill set to handle and analyze high-dimensional omics data.
Figure 2: App-based web interface
PanHunter® is structured into apps, each with a specific focus, covering a broad spectrum of different analysis tasks. All this is built on top of a curated set of published and peer-reviewed, or in-house developed, robust algorithms.
To empower our scientists to cope with the overwhelmingly increasing amount of data and to reduce the complexity of data analysis and interpretation, we built PanHunter, our versatile and interactive multi-omics data analysis platform. It provides a plethora of tools, visualizations, and reference information readily available. Via its easily accessible web interface, it allows our scientists to immerse themselves in the data, analyse them faster and more focused, visualize them easier, put them into context, and derive deep and meaningful interpretations from them to make well-informed decisions. PanHunter can be used through its graphical user interface in any modern web browser. This interactive interface provides immediate feedback to user interaction and reduces waiting times between setting up and interpreting analyses, especially compared to the classical pipelining approach.
This allows our users to focus on the results rather than the process of handling the data. Additionally, the modular structure of PanHunter allows quick and easy adaption to new tasks and necessity arising from our internal research projects or external partner requests.
The main motivation when designing PanHunter was to reduce existing obstacles for data access as much as possible and diminish the overhead and repetitive work usually associated with omics data analysis. The result is that both, our bioinformaticians, as well as our lab scientists can focus more on data interpretation and less on data processing. However, using one general platform for omics data analysis comes with additional advantages: standardization, reproducibility, and collaboration.
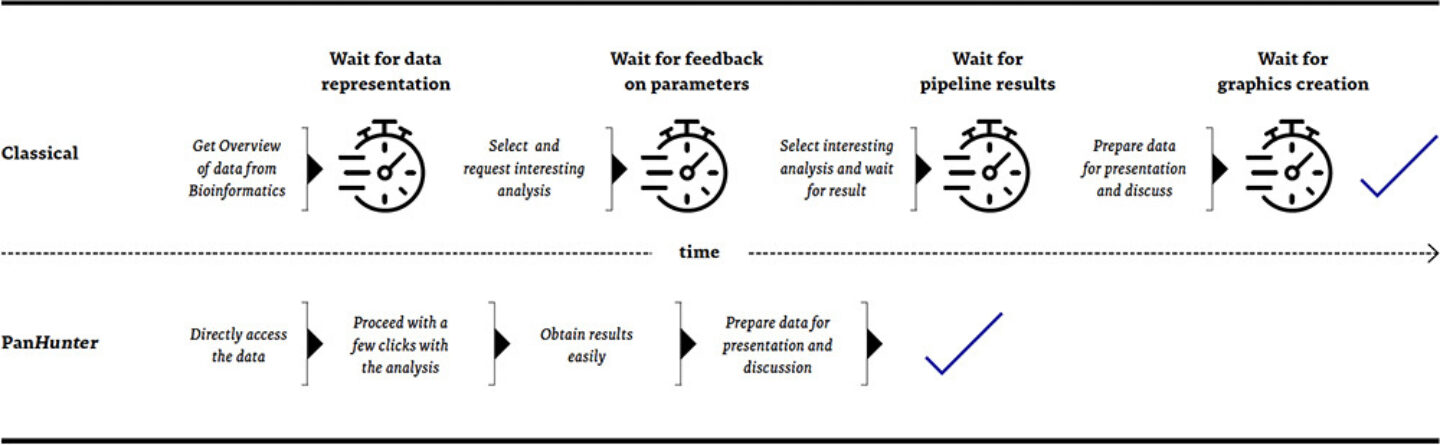
Figure3: Comparison of a biologist’s journey from data presentation towards result generation
A standardized data processing workflow, which is an integral part of PanHunter, allows for easy re-purposing of already existing datasets in other contexts, which eventually increases the value of each individual experiment. It serves therefore as an exchange and access platform for the increasing number of linkable datasets at Evotec and its partners. Furthermore, our software generates reproducible and comparable results, since all intermediate steps, algorithms, and parameters are tracked automatically. And finally, all this is fully equipped for global collaboration of many scientists in larger projects, as simultaneous access to the same data base and immediate sharing of generated results is build right into the core of PanHunter.
After all, omics data alone only provide one part of the picture. To unfold their full potential, our software provides everything that is necessary to put the omics data into perspective by accessing additional meta information, reference data, chemical and structural information for compounds, and even clinical information for projects containing human samples.
In most situations, this vast amount of additional information is automatically associated with the experimental data. This allows the user to easily pull, e.g., gene or protein information from associated databases or run statistical tests to identify metabolic pathways or protein interaction networks that are significantly regulated within the tested data. Such networks can reveal the underlying molecular mechanisms for a disease or drug treatment.
Whenever performing such statistical methods, PanHunter applies well-selected default parameters, while being always transparent about them and providing the option to customize. This allows us to serve all scientists from a broad experience spectrum: new users to the field of omics data analysis will be guided and can rely on the default settings while experienced users can customize almost any small details of an analysis.
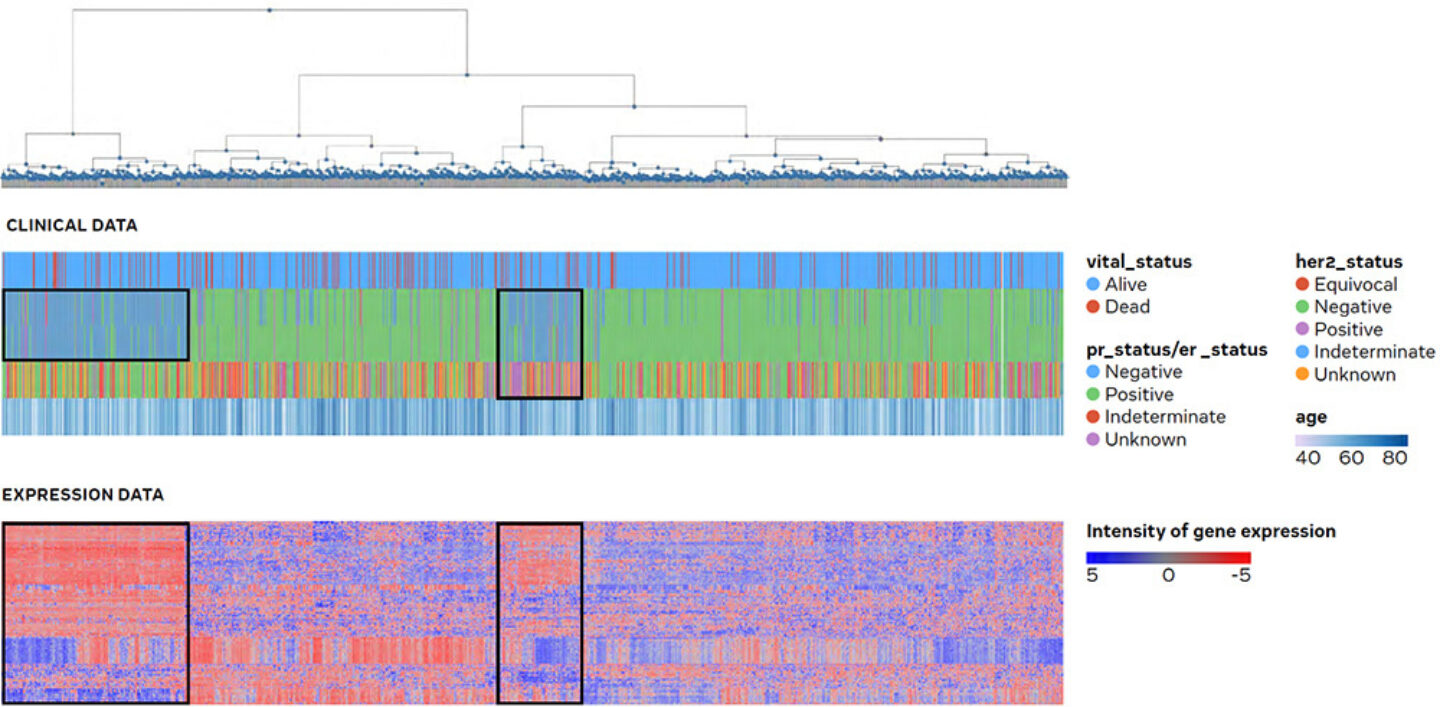
Figure 4: Global view and correlation of clinical vs. transcriptomic data: tumour tissue samples from the public breast cancer cohort of The Cancer Genome Atlas (TCGA) are shown in PanHunter and allow to associate gene expression with clinical parameters. Hereby the user is able to identify potential connections in a global view on gene expression.
While the user gets empowered to perform a plethora of different analysis via the GUI of PanHunter that requires no coding or scripting skills, all other aspects, like managing the data and carrying out the analysis processes, are handled in the background. The omics data are stored in a fast yet secure data layer. In addition to the experimental and meta data provided by the user, this data layer handles and provides access to all additional information already mentioned above.
The processing, analysis, and interpretation of omics data involves a series of sequential steps, all of which are covered by PanHunter. As soon as new omics data are generated, e.g., via mRNA library sequencing using next-generation sequencing (NGS), the data needs to be pre-processed to generate a so-called feature abundance table.

“Omics-driven drug discovery is the cornerstone of disease understanding on the molecular level and the fast lane to cures!”
Cord Dohrmann, CSO Evotec SE
In case of RNA-Seq, this table contains for all samples the abundance/ activity information for all genes. For other omics types, e.g., the genotype for all single nucleotide polymorphisms (SNPs) of all individuals is determined (genomics), the relative abundance of proteins for all samples is quantified (proteomics), or the amount of certain metabolites is measured (metabolomics). For all these quantification steps, PanHunter has its own, versatile, and robust pre-processing pipeline that can ingest the raw data and information from a variety of locations. Additionally, the software provides the necessary assistance tools to the user, to supplement the omics data with the experimental meta data (e.g., which tissue type, treatment, or time point belongs to which sample).
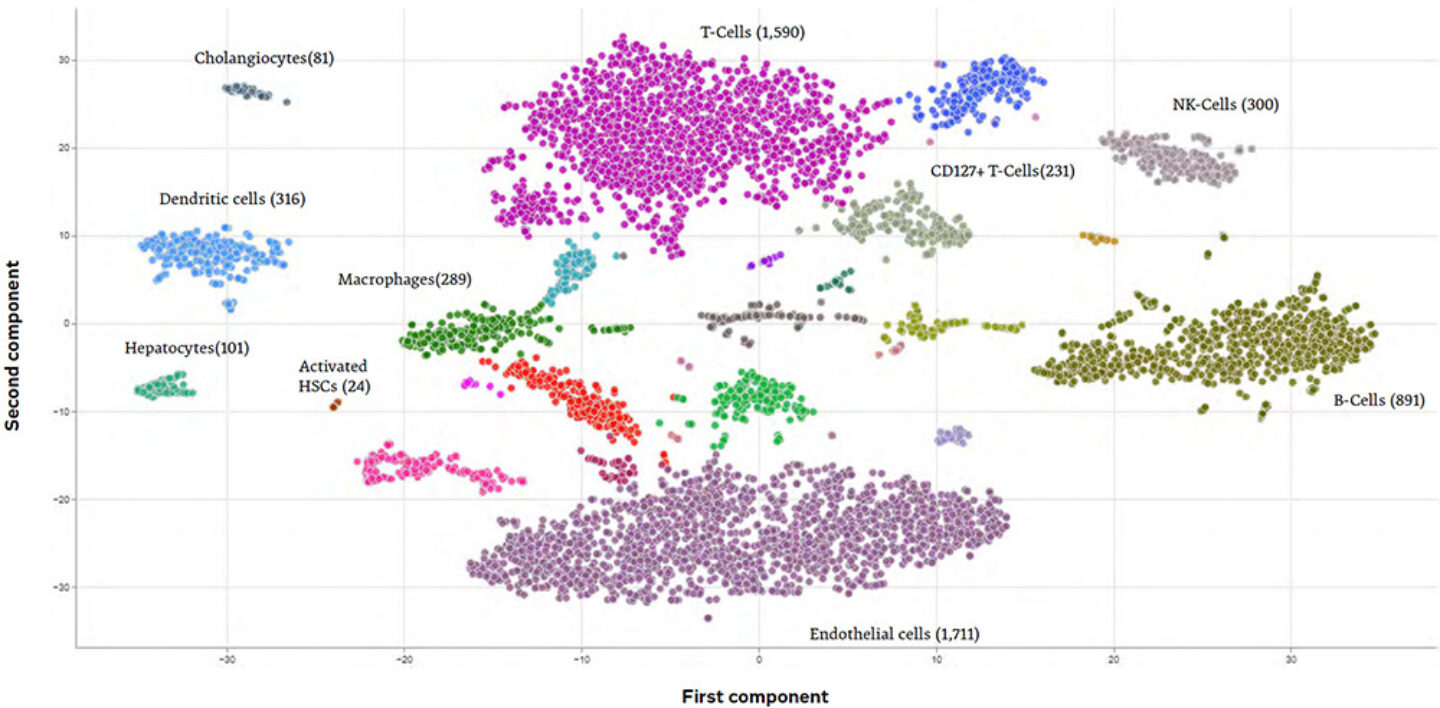
Figure 5: Illustration of automated AI-driven cell type annotation for clusters of liver scRNAseq cells
Once all this information is integrated and available within PanHunter, users can use them for differential analysis, to identify significantly regulated features (e.g., up- or downregulated genes, proteins, or metabolites), or statistical correlation testing, for example when trying to identify molecular marker genes based on the abundance of certain clinical parameters or survival rates. Moreover, PanHunter offers dedicated tools for specific omics data types, like single-cell or spatial transcriptomics. For example, it is easily possible for single-cell datasets to perform the key tasks of cell type clustering and annotation, with or without the assistance of machine-learning algorithms, or for spatial transcriptomics to associate a given cell’s gene activity information with the exact location on its originating histological slice. After all, PanHunter became the central platform for interdisciplinary omics data analysis in a large variety of internal and external projects and keeps growing constantly.

Figure 6: A cross-omics analysis of samples linked by meta-data


