Optimize membrane protein solubilization screening with the efficient ALIAS autosampler setup.
Introduction
Membrane proteins such as G protein-coupled receptors (GPCRs), ion channels, and transporters are involved in essential cellular processes and their location on cell surfaces allows them to be easily accessible for drug molecules making them attractive drug targets. Recent developments in structural biology (mainly cryo-electron microscopy) have made studying membrane proteins much more feasible. However, purification of membrane proteins presents different challenges compared to soluble proteins. Membrane proteins are usually expressed in lower amounts and are less stable than soluble proteins. Furthermore, within cell membranes these proteins are embedded within a lipid bilayer making them insoluble in aqueous buffers. Therefore, before purifying membrane proteins, they need to be extracted into suitable detergents and lipid combinations to stabilize them. There has been an enormous growth in the field of membrane protein extraction in the last decade with further developments using amphipols and nanodiscs. Every new membrane protein target presents a new challenge, and the solution could be novel. This presents a challenge to screening various extraction methods for membrane proteins. In this report, we will only focus on screening for detergent and lipid combinations for the extraction of membrane proteins. The method explained here can be used for other extraction strategies, too.
This article presents a method to efficiently screen for optimal detergent conditions for membrane protein solubilization using an analytical size-exclusion chromatography (aSEC)/fluorescence-detection size-exclusion chromatography (fSEC) setup enhanced by an ALIAS autosampler, allowing for rapid testing of 48 conditions per day.
Instrumentation setup of analytical size-exclusion column with UV and fluorescence detection (aSEC/fSEC)
Small volume and high-resolution analytical size-exclusion columns have been routinely used to assess the hydrodynamic radii and apparent molecular weights of macromolecules . Columns are readily available with various base matrices (Superdex™, Superose™, Sephacryl™, etc.) and pore sizes to enable separation of macromolecules of different size ranges . Since membrane protein expression generally has lower yields, standard UV detection is usually not sufficient to detect the eluted peak and hence fluorescence detection can be implemented as an alternative. Routinely, recombinant membrane proteins are expressed with a fluorescent tag (e.g., GFP), or the tag is added post-production by specifically targeting His tags. In the process of analyzing the products, cell pellets in our lab are subjected to an hour of solubilization before analysis. In the workflow described below (Figure 1) the clarified solubilized lysate is injected using an ALIAS autosampler in conjunction with an ÄKTA pure™ system attached to analytical size-exclusion chromatography (aSEC)/fluorescence-detection size-exclusion chromatography (fSEC) ÄKTA pure™ system. The use of an ALIAS autosampler in combination with a standard aSEC/fSEC setup increases throughput and process efficiency as well as enabling analysis of low volume samples.
The setup has also been tested to function within a 4 °C cabinet or cold room.

Figure 1: A standard instrumentation setup for an analytical sizing column with UV and fluorescence detection
Detergent optimization screen
A standard detergent screen is performed using 0.4 L culture. Pellets are harvested in 10 mL aliquots. After lysis, each 10 mL culture is first subjected to a low-speed centrifugation to remove cell debris. Following this, a high-speed centrifugation is performed to pellet the membranes (if the membrane protein abundance is low, omit this step, and instead use whole cell solubilization). Membrane pellets are solubilized in high concentration detergent (typically 1%) for 45 – 90 mins at 4 °C (see Table 1). (The length of solubilization time could be optimized by exploring this variable with a secondary screen after identifying the most promising detergent). Ultracentrifugation is used to separate solubilized membrane proteins. If membrane proteins are not expressed with a fluorescent tag, a His/Avi tag specific fluorophore is added to the lysate and the samples are injected onto the size-exclusion column. The remaining lysate was subjected to automated affinity purifications and the eluted peaks were further used for aSEC/fSEC analysis. This step helps to confirm the findings from fSEC analysis.
aSEC = analytical size-exclusion chromatography; CMC= critical micellar concentration; fSEC = fluorescence-detection size-exclusion chromatography; ND = not determined; DDM = n-Dodecyl-β-D-Maltopyranoside; GDN = Glyco-Diosgenin; LMNG = Lauryl Maltose Neopentyl Glycol; OG = Octyl-β-D-Glucopyranoside; DM = n-Decyl- β-D-Maltoside.
A typical detergent screen could include 24–48 different conditions (Table 1). This number substantially increases when applied to multiple constructs of a target (domain boundaries, homologues, stabilizing mutations, etc.). The ALIAS autosampler allows injection of low volume samples from a standard 96/384-well plate or vials. The example below (Figure 2) shows a result from a solubility screening of a target membrane protein, which was successfully isolated with detergent and lipid optimization.
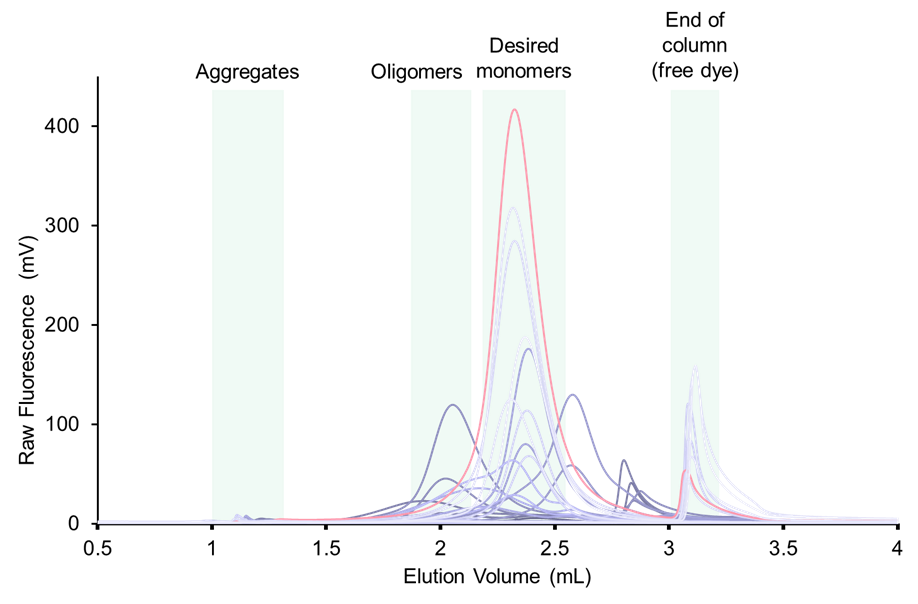
Figure 2: The above image shows an overlay of 36 fluorescence traces from a S200 increase 5/150 column. The best solubilization detergent and lipid mixture is shown in orange. Each run is performed in a buffer containing varying amounts of detergent/lipid combinations (captured in Table 1)
Summary and conclusion
Membrane proteins are essential targets in drug discovery. There are many new methods to enable successful extraction. This article briefly presents a way to screen for optimal detergent conditions. Our aSEC/fSEC setup enables us to successfully determine the fraction of membrane proteins solubilized as a non-aggregated fraction. The addition of an ALIAS autosampler to this setup enables us to screen 48 conditions in a day in the cold room. This methodology, with an increase in process efficiency and productivity, describes a possible routine method to screen for membrane protein solubilization.
Cytiva and the Drop logo are trademarks of Life Sciences IP Holdings Corporation or an affiliate doing business as Cytiva. ÄKTA pure, Sephacryl, Superdex, and Superose are trademarks of Global Life Sciences Solutions USA LLC or an affiliate doing business as Cytiva. Any other trademarks are the property of their respective owners.
Interested in learning more?
